Environment
The framework is grid based and each environmental variable is represented by a seperate raster layer.
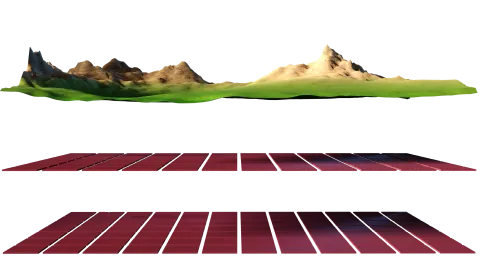
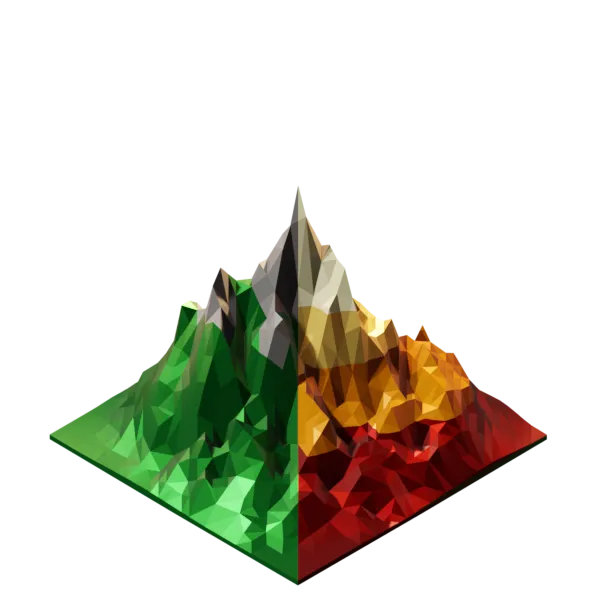

Environment
The framework is grid based and each environmental variable is represented by a seperate raster layer.

Species
This environment is inhabited by one or multiple species, where each population is represented by one grid cell of the environment.

Mechanisms
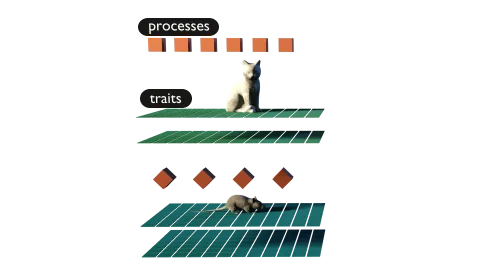
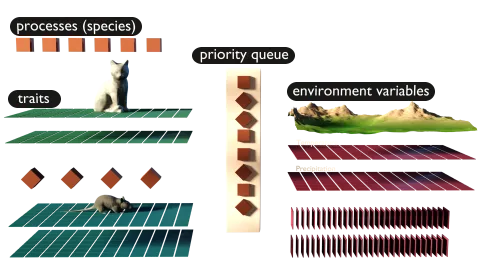
Species are defined by traits (any type of biological meaningful data) and processes (mechanisms that define how the species interacts with its environment and other species).
Execution
The processes are sorted into a queue, based on a user given priority, and executed each time step.
You can install the R package from CRAN with:
install.packages("metaRange")Or view and install it from the GitHub repository for the latest development version.
devtools::install_github("metaRange/metaRange")Fallert, S., Li, L., & Cabral, J. S. (2025). metaRange: A framework to build mechanistic range models. Methods in Ecology and Evolution, 16, 49–56. https://doi.org/10.1111/2041-210X.14461